Abstract
Inherited and acquired bone marrow failure syndromes (BMF) may be difficult to distinguish due to heterogeneity and overlap of clinical phenotypes. Genomic screening has been increasingly used to identify mutations in BMF-related genes that are known to be etiologic in inherited BMF. However, genomic testing is expensive, results may not return for several seeks, and findings can be difficult to interpret as some reported variants are of unclear clinical significance. To guide the decision-making for genetic testing and results interpretation, we aimed to identify clinical and molecular parameters associated with a higher probability of patients having an inherited disease. We screened 323 BMF patients from two independent cohorts for germline mutations in BMF-related genes using a targeted next-generation sequencing (NGS) assay, and correlated the results with patients' prior diagnosis, family history, telomere length (TL), karyotype, and the presence of a paroxysmal nocturnal hemoglobinuria (PNH) clones.
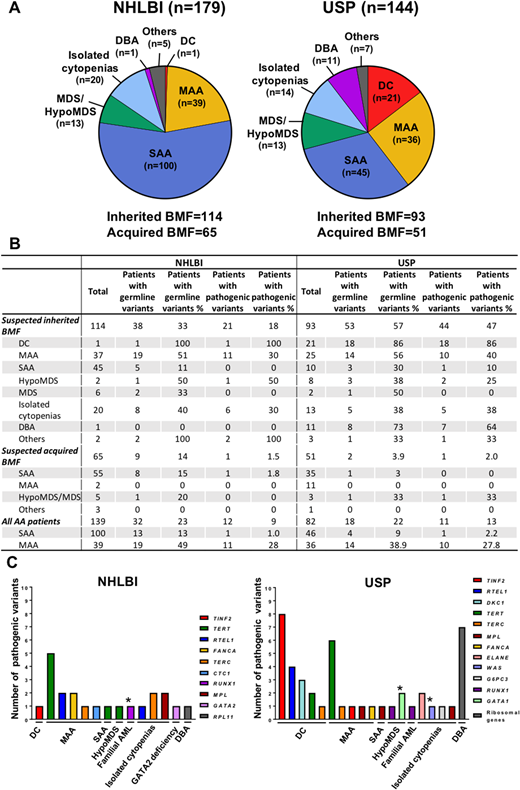
Patients were followed at the Hematology Branch of NHLBI (NHLBI, n=179) and the Ribeirão Preto Medical School, University of São Paulo (USP, n=144). Diagnoses included were severe (SAA) and moderate aplastic anemia (MAA), isolated cytopenias, myelodysplastic syndrome (MDS), hypocellular MDS (HypoMDS), dyskeratosis congenita (DC), and Diamond-Blackfan anemia (DBA). Patients were classified as suspected to have inherited BMF (phenotype suggestive for constitutional disease, short or very short telomeres, family history of hematologic, pulmonary, or liver disease, and idiopathic cytopenias), or acquired BMF (normal TL and no signs of constitutional disease) (Figure 1A). Pathogenicity of novel and rare variants was assessed using the ACMG criteria.
We identified a pathogenic (or likely pathogenic) germline variant in 21 (18%) and 44 (47%) inherited BMF patients from NHLBI and USP cohorts, respectively (Figure 1B). Altogether, mutated genes were associated with telomeropathies (mostly DC and MAA), congenital cytopenias, DBA, cryptic Fanconi anemia, and myeloid malignancies (Figure 1C). In both cohorts, inherited BMF patients with DC, DBA, MAA, and isolated cytopenias were more likely to have a pathogenic variant. BMF patients suspected to have an acquired disease were rarely found with a pathogenic variant; one patient from each cohort (NHLBI, 1.5% and USP, 2%), carried the R166A RUNX1 and A202T TERT variants, respectively. Overall, patients with SAA were highly unlikely to have a pathogenic variant, regardless of the clinical suspicion for constitutional disease (Figure 1B). The presence of PNH clone and chromosomal abnormalities were poorly associated with variants' pathogenicity; only one patient from the USP cohort had a PNH clone of 6% and the pathogenic TERT D718E variant, and three patients had an abnormal karyotype (indicated by asterisks in Figure 3C). In both cohorts, we additionally screened 101 acquired BMF and 140 inherited BMF patients for somatic clones in myeloid-driver genes. These results recapitulated the clonal landscape previously observed in AA by our group; the frequency of variants in ASXL1, DNMT3A, TET2, and JAK2, but not in BCOR and BCORL1, increased with aging. In the current study, TP53, RUNX1, and Ras genes were more frequently mutated in the patients suspected to have inherited BMF (Fisher's exact test, 14% vs. 4.4%; p<0.005) whereas BCOR and BCORL1 were more commonly abnormal in patients suspected to have acquired BMF (18% vs. 3.1%; p<0.005). Somatic mutations were particularly present in 5/21 DC patients (23%, median age, 11 years) but not in DBA (1 out of 11; median age, 3) and isolated cytopenias (median age, 6).
In summary, inherited BMF patients were more likely to have a pathogenic variant compared to acquired BMF (18% vs. 1.5%, p<0.001). Inherited BMF patients with MAA and isolated cytopenias without PNH clones and a normal karyotype had increased risk of having constitutional disease. Systematic analysis of clinical and genomic data may be helpful to assist physicians in identifying patients who should be first screened for inherited BMF based on the probability of finding a pathogenic germline variant.
Dunbar:National Institute of Health: Research Funding. Young:CRADA with Novartis: Research Funding; GlaxoSmithKline: Research Funding; National Institute of Health: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal